Submit a Review & Earn an Amazon Gift Card
You can now submit reviews for your favorite Tocris products. Your review will help other researchers decide on the best products for their research. Why not submit a review today?!
Submit ReviewMPD 2 is a SARS-CoV-2 Mpro Degrader (PROTAC®) (DC50 = 296 nM in 293T cells). Mpro degradation is time-dependent and CRBN-mediated. Antiviral in A549-ACE2 cells infected with several SARS-CoV-2 strains (EC50 = 492 nM at delta variant) and against a nirmatrelvir-resistant mutant of SARS-CoV-2, NSP5 E166A (EC50 = 460 nM).
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
 View Larger
View Larger
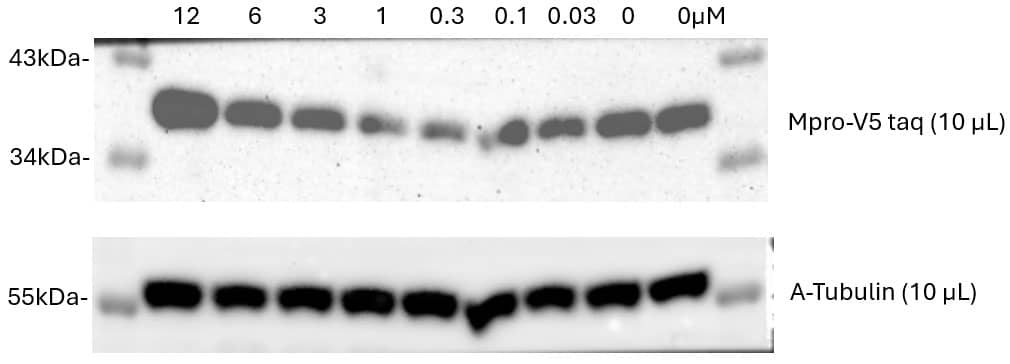
Application of MPD 2. Western blot data showing dose responsive degradation with MPD 2 on transiently expressed SARS-CoV-2 Main Protease in HEK293 cells. 1st Ab: HRP Anti-V5 tag antibody [E10/V4RR] (ab173837) (1:2000), 2nd Ab: Anti-Mouse HRP (1:5000). Loading control: 1st Ab: Anti-alpha - Tubulin antibody [DM1A] - (1:10000) 2nd Ab: Anti-mouse HRP (1:5000). Data provided by Brönstrup Lab at the Helmholtz Centre for Infection Research.
| 分子量 | 973.13 |
| 公式 | C51H68N6O13 |
| 储存 | Store at -20°C |
| PubChem ID | 171663340 |
| InChI Key | ZKSDAFLRLHGTNH-DTKLLKMQSA-N |
| Smiles | O=C1[C@@H](CCN1)C[C@H](NC([C@@H](NC([C@@H](NC(OCC2=CC(OCCCCCCOC3=C4C(C(N(C4=O)C5CCC(NC5=O)=O)=O)=CC=C3)=CC=C2)=O)[C@@H](C)OC(C)(C)C)=O)CC6CCCCC6)=O)C([H])=O |
上方提供的技术数据仅供参考。批次相关数据请参见分析证书。
Tocris products are intended for laboratory research use only, unless stated otherwise.
| 溶剂 | 最高浓度 mg/mL | 最高浓度 mM | |
|---|---|---|---|
| 溶解性 | |||
| DMSO | 97.31 | 100 |
以下数据基于产品分子量 973.13。 Batch specific molecular weights may vary from batch to batch due to the degree of hydration, which will affect the solvent volumes required to prepare stock solutions.
| 浓度/溶剂体积/质量 | 1 mg | 5 mg | 10 mg |
|---|---|---|---|
| 1 mM | 1.03 mL | 5.14 mL | 10.28 mL |
| 5 mM | 0.21 mL | 1.03 mL | 2.06 mL |
| 10 mM | 0.1 mL | 0.51 mL | 1.03 mL |
| 50 mM | 0.02 mL | 0.1 mL | 0.21 mL |
参考文献是支持产品生物活性的出版物。
Alugubelli et al (2024) Discovery of first-in-class PROTAC Degraders of SARS-CoV-2 main protease. J.Med.Chem. 67 6495 PMID: 38608245
If you know of a relevant reference for MPD 2, please let us know.
关键词: MPD 2, MPD 2 supplier, MPD2, degrader, degraders, degrade, degrades, Mpro, main, protease, SARS-CoV-2, sars, cov, protac, proteolysis, targeting, chimera, CRBN, cereblon, e3, ligase, Other, Degraders, 3C, and, 3CL, Proteases, 8802, Tocris Bioscience
目前没有该产品的评论。 Be the first to review MPD 2 and earn rewards!
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image
Tocris offers the following scientific literature in this area to showcase our products. We invite you to request* your copy today!
*请注意,Tocris 仅会向正规科研企业/机构地址发送文献。
This brochure highlights the tools and services available from Bio-Techne to support your Targeted Protein Degradation and Induced Proximity research, including:
Degraders (e.g. PROTACs) are bifunctional small molecules, that harness the Ubiquitin Proteasome System (UPS) to selectively degrade target proteins within cells. They consist of three covalently linked components: an E3 ubiquitin ligase ligand, a linker and a ligand for the target protein of interest. Authored in-house, this poster outlines the generation of a toolbox of building blocks for the development of Degraders. The characteristics and selection of each of these components are discussed. Presented at EFMC 2018, Ljubljana, Slovenia