Fluorescent Probes for Imaging Bacteria
Fluorescent probes for imaging bacteria are useful for advancing bacterial research and antibiotic design. One technique for studying bacteria, is to incorporate fluorescent probes such as Fluorescent D-amino acids (FDAAs) into the cell wall of live bacteria. FDAAs are suitable for use with confocal and super-resolution microscopy (SRM).
| Cat. No. | Product Name / Activity |
|---|---|
| 7973 | 5-InoAz |
| Metabolic probe for labeling mycobacterium cell envelope | |
| 6798 | Click N-Acetylmuramic acid - alkyne |
| Bacterial peptidoglycan derivative; suitable for 'click'-conjugation to fluorescent dyes | |
| 7506 | Click N-Acetylmuramic acid - azide |
| Bacterial peptidoglycan derivative; suitable for 'click'-conjugation to fluorescent dyes | |
| 7714 | EDA-DA |
| Unnatural dipeptide building block with alkyne group for functionalizing peptidoglycan | |
| 6647 | HADA |
| Blue fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 6648 | NADA-green |
| Fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 7408 | OGDA |
| Green fluorescent D-amino acid; compatible with STED microscopy | |
| 6649 | RADA |
| Orange-red TAMRA-based fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 7406 | Rf470DL |
| Blue rotor-fluorogenic fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 8013 | RMR-Tre |
| Far-red fluorogenic trehalose probe for live mycobacteria imaging | |
| 7860 | sBADA |
| Green fluorescent D-amino acid for labeling peptidoglycans in bacteria | |
| 7834 | sCy5DA |
| FDAA for super-resolution microscopy of bacteria | |
| 7835 | sCy5DL-amide |
| FDAA for super-resolution microscopy of bacteria | |
| 7449 | Se-NADA |
| Orange fluorescent benzoselenadiazole D-amino acid (FDAA) for imaging bacteria; also photosensitizer | |
| 6802 | 6 TMR Tre |
| Fluorescent trehalose; selectively labels mycobacterial cell envelope | |
| 6650 | YADA |
| Green-yellow lucifer yellow-based fluorescent D-amino acid for labeling peptidoglycans in live bacteria |
Antibiotic Research: Targeting peptidoglycans (PG)
The investigation of bacterial physiology is essential for developing antibiotics to treat diseases such as E. coli infection. Targeting PG biosynthesis is a common approach to designing antibiotics, since it is essential for bacterial growth and function, as well as being selectively lethal to bacteria without harming human cells.
PGs are components of bacterial cell wall, which protect the bacteria from environmental stresses. In most bacteria, a peptidoglycan cell wall is made up of macromolecular polymers of glycan strands that link to the cytoplasmic membrane via oligopeptide bridges.
Bacteria are split into two groups, gram-negative and gram-positive, depending on the organization of the PGs. Gram-negative bacteria have a PG layer of about 5 nm, between a cytoplasmic membrane and the outer membrane; gram-positive bacteria have a much thicker PG layer of about 20-50 nm on the periplasmic side of the cytoplasmic membrane. Mycobacteria do not fit in to either category, they have a PG layer and a waxy surface layer composed of mycolic acids, lipids and arabinogalactan.
The ability to investigate PG physiology in high resolution is a useful development for bacterial research and antibiotic design. New SRM techniques are enabling researchers to study bacteria in high resolution under physiologically relevant conditions to identify new therapeutic vulnerabilities.
Techniques for Imaging and Staining Bacteria
Previous techniques used for observing bacteria include electron microscopy, but this method had the disadvantage of using mercury-containing chemicals. The fixation and staining required for this technique precluded work with live bacteria. The next advancements came from fluorescent microscopy which has been useful for gaining a greater understanding of bacterial physiology and function in live cells. However, this technique is limited because spatial resolution is restricted by the diffraction limit of incident light to ~250 nm (Abbe diffraction limit). Recent advances of both super-resolution microscopy techniques (including STORM, SIM, STED and PALM) and new molecular probe types (namely Fluorescent D-amino acids (FDAAs)) has accelerated the understanding of peptidoglycan biosynthesis dynamics and mechanisms, allowing unprecedented analysis at a molecular level.
Using FDAAs to Stain Bacteria
Previously fluorescently-labeled antibiotics, and fluorescently-labeled wheat germ agglutinin (FWGA) were used as molecular probes to study PGs. Both of these probes have major drawbacks, fluorescent antibiotics suppress bacterial growth, and FWGA cannot permeate the cell walls of gram-negative bacteria.
FDAAs are metabolic probes that leverage the promiscuous nature of the bacterial PG 60 biosynthetic pathway. FDAAs are small molecules conjugated to D-amino acids and are incorporated into new poly-glycan chains. They efficiently label peptidoglycans in bacterial cell walls in situ, allowing the study of bacteria morphology and formation, as well as bacterial growth. FDAAs can be used in gram-negative and gram-positive bacterial species, in real-time and in live cells, with minimal toxic effects. They are suitable for use with both confocal and super-resolution microscopy.
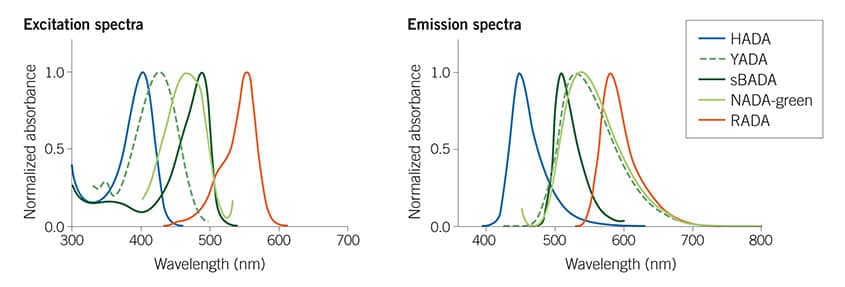
Figure 1: The Tocris Fluorescent D-amino acids (FDAAs) collection spans the visible spectrum, giving useful multiplexing options. For more details read Hsu et al (2017) Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem.Sci. 8 6313 PMID: 28989665
5 star FDAA review - featuring HADA (Cat. No. 6647)
