Unnatural Amino Acids / Non Canonical Amino Acids
Unnatural Amino Acids (UAAs) or Non-Canonical Amino Acids can be incorporated into proteins at defined positions. Unnatural amino acids can be used to investigate the structure and dynamics of proteins, to study their interactions and functions, to control their activity in living cells, as well as to introduce novel functions in proteins unachievable in nature.
| Cat. No. | Product Name / Activity |
|---|---|
| 5113 | AbK |
| Photo-crosslinking probe; unnatural amino acid | |
| 7936 | Ac-Cys-NHMe |
| Building block for the lysine acylation using conjugating enzymes (LACE) technique | |
| 8086 | Acridonylalanine New |
| Fluorescent unnatural amino acid. | |
| 7370 | 3-Aminotyrosine |
| Unnatural amino acid for inducing red shift in fluorescent proteins and fluorescent protein-based biosensors | |
| 6584 | L-Azidohomoalanine hydrochloride |
| Unnatural amino acid for bio-orthogonal labeling of newly synthesized proteins | |
| 6585 | L-Azidonorleucine hydrochloride |
| Unnatural amino acid for bio-orthogonal or fluorescent labeling of newly synthesized proteins | |
| 6315 | DMNB-caged-Serine |
| Caged serine; excited by visible blue light | |
| 7714 | EDA-DA |
| Unnatural dipeptide building block with alkyne group for functionalizing peptidoglycan | |
| 6647 | HADA |
| Blue fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 6648 | NADA-green |
| Fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 7361 | ProK |
| Unnatural amino acid, propargyl lysine derivative | |
| 6649 | RADA |
| Orange-red TAMRA-based fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 7406 | Rf470DL |
| Blue rotor-fluorogenic fluorescent D-amino acid for labeling peptidoglycans in live bacteria | |
| 6650 | YADA |
| Green-yellow lucifer yellow-based fluorescent D-amino acid for labeling peptidoglycans in live bacteria |
What are Unnatural Amino Acids / Non-Canonical Amino Acids?
An amino acid is as an organic compound which possesses an amine (-NH2) and a carboxylic acid (-COOH) functional group. There are 20 naturally occurring amino acids, which vary only by their side chain, or R-group. These are genetically encoded in all living organisms and form the building blocks of proteins.
Unnatural amino acids or non-canonical amino acids are non-proteinogenic amino acids that are either found naturally in organisms or are synthetically made in a laboratory. Crucially they are amino acids that are not located in the genetic code of naturally occurring organisms. There are however, over 100 known "unnatural" amino acids occurring in nature as either intermediates in biosynthesis pathways or in proteins following post-translational modifications. They have been found to play roles as neurotransmitters and toxins, amongst many other functions.
Synthetically made unnatural amino acids have been engineered to contain a variety of artificial side chains. These non-natural side chains allow for versatile uses in biological studies and can be incorporated into cellular proteins either site-specifically, via genetic code expansion or via residue-specific incorporation.
Applications of Unnatural Amnio Acids / Non-Canonical Amino Acids
Unnatural amino acids are useful in many areas of research. For example, they can be incorporated into fluorescent proteins to alter spectral properties (3-Aminotyrosine Cat. No. 7370), form cross-links to identify native protein-protein interactions (Dizpk Cat. No. 7284 coming soon) as well as being used in FRET for structural and mechanistic studies (L-ANAP Cat. No. 7359 coming soon). Unnatural amino acids can also be employed to analyze protein synthesis using the techniques bio-orthogonal non-canonical amino acid tagging (BONCAT) or fluorescent non-canonical amino acid tagging (FUNCAT). An appropriately tagged unnatural amino acid can access bio-orthogonal chemistries to achieve specific labeling in live cells, which can be a useful way to selectively incorporate fluorescent probes for live cell imaging. Another function is to incorporate them into enzymes to explore, modulate or improve their activity. Other unnatural amino acids are photoreactive, such that exposure to light of a specific wavelength triggers the formation of covalent crosslinks, enabling selective and potent inactivation or activation of protein function. Photoactivatable unnatural amino acids can be used to investigate the function of proteins such as ion channels, transporters and GPCRs.
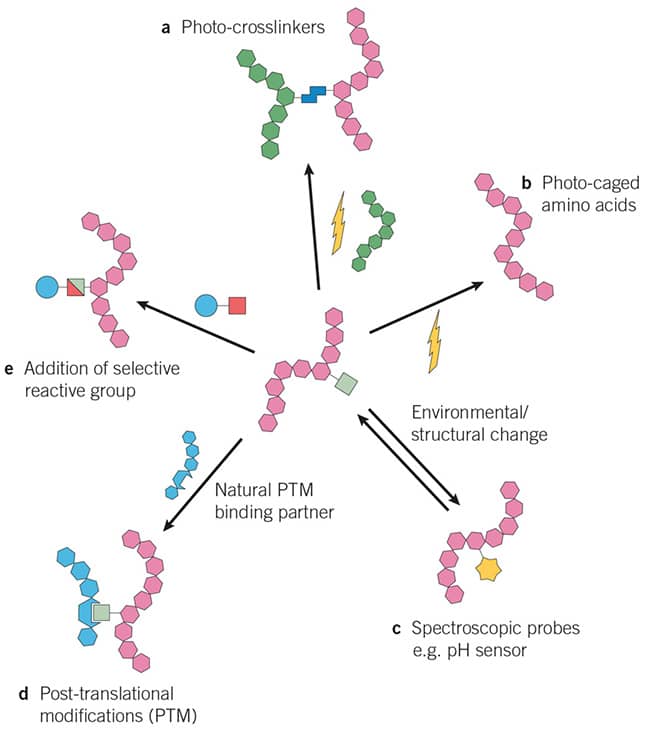
Figure 1: Main types of genetically encoded unnatural amnio acids / non-canonical amino acids tools. A: Photo-crosslinkers, b: Photo-caged amino acids, c: Spectroscopic probes e.g. pH sensor, d: Post-translational modifications (PTM), e: Addition of selective reactive group e.g. used to investigate biological pathways.
Image adapted from Dumas et al. (2015) Designing logical codon reassignment - Expanding the chemistry in biology. Chem Sci. 6 50. PMID 28553457
What is Genetic code expansion?
One method for incorporating unnatural amino acids into proteins is by genetic code expansion. This is a widely used method in synthetic biology, protein engineering and structural and functional studies. It involves repurposing key parts of a cell's translation machinery (tRNA and aminoacyl-tRNA synthetase) to incorporate an unnatural amino acid site selectively within a protein.
This method requires:
- A source of the unnatural amino acids to encode (e.g. added to growth media),
- An unused codon to adopt (usually a STOP codon),
- An orthogonal tRNA that recognizes the adopted codon,
- An orthogonal aminoacyl-tRNA synthetase (aaRS) that recognizes the orthogonal tRNA and can charge it with the desired unnatural amino acid.
*Note: the orthogonal aaRS should only acylate (charge) the orthogonal tRNA with the designated unnatural amino acid to ensure site selective incorporation into the protein. Conversely orthogonal tRNA and unnatural amino acids should not be substrates of the endogenous aminoacyl-tRNA synthetases.
A cell's translation machinery translates every nucleotide triplet codon on mRNA into the corresponding amino acid. Endogenous aminoacyl-tRNA synthetase (aaRS)/tRNA pairs translate 61 of the 64 codons into the 20 canonical amino acids. The other three codons (UAG, UGA and UAA), are stop codons, which terminate the translation process. In genetic code expansion these can instead be repurposed for use with the orthogonal parts of the translation machinery. The most frequently used codon for this purpose is the UAG "amber stop codon" due to its infrequent occurrence in nature. In addition, some genetic code expansion methods have used four nucleotide codons as blank codons, enabling multiple unnatural amino acids to be incorporated simultaneously. This leads to lower translation efficiency in mammalian cells but can be effectively used in Escherichia coli expression systems.
One of the most used genetic code expansion pairs are the Pyrrolysyl-tRNA synthetase (PylRS)/tRNAPyl pairs derived from Methanosarcina barkeri (Mb) and Methanosarcina mazei (Mm). This is because this pair are orthogonal in the prokaryote Escherichia coli system and mammalian systems, and fortuitously, tRNAPyl recognizes the UAG codons, so removing the need for an engineered tRNA. The diversity of available aaRS/tRNA pairs means that multiple unnatural amnio acids can be incorporated at the same time into the same protein, allowing different combinations of activity to be investigated.
Genetic code expansion allows investigation of modified proteins within its native environment under physiological conditions, which is especially important when you consider that a protein's function can be dependent on its microenvironment or cell cycle stage. Introduction of unnatural amino acids by genetic code expansion can be used to modulate a target, causing it to become permanently active, inactivated or giving it a reversible functionality.
Genetic Code Expansion: Translational Control of Protein Function by Amber Suppression
This method of control involves the insertion of an amber stop codon into a target gene by DNA mutagenesis. For incorporation of the unnatural amino acid into the protein of interest, the appropriate unnatural amino acid is added to the reporter system (e.g. in the cell's growth media) enabling the orthogonal tRNA to become acylated. Now charged with the unnatural amino acid, the tRNA can attach at the appropriate tRNA binding site on the ribosome and translate the amber codon, incorporating the unnatural amino acid into the protein chain. This process is known as amber suppression and is the most commonly used method for genetic code expansion. By removing the unnatural amino acid from system (e.g. growth media switched) protein translation will halt at the amber stop codon, creating a non-functional protein.
Translational control is particularly useful in cellular reporter systems that rely on the expression or suppression of fluorescent proteins, RNA aptamers or luciferases. This system has also been employed in CRISPR gene editing, allowing the user temporal control of gene editing in a particular system.
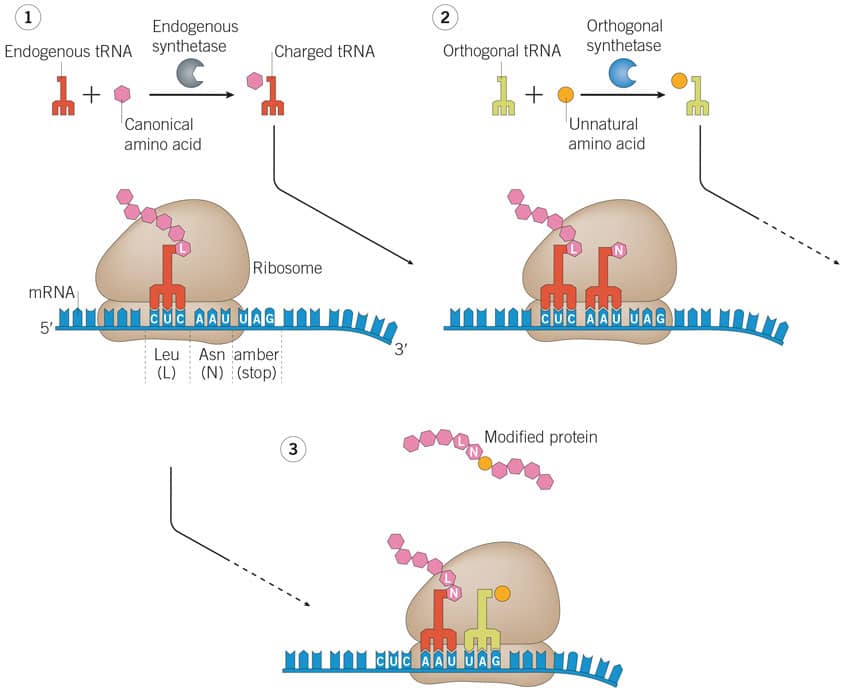
Figure 2: Genetic code expansion adding unnatural amnio acids by translational control of protein function using amber suppression. Image adapted from Nodling, et al (2019) Using genetically incorporated unnatural amino acids to control protein functions in mammalian cells . Essays Biochem. 63 237 PMID 31092687
Genetic Code Expansion: Optical Control of Protein Function by Unnatural Amino Acids
Using light to modify unnatural amino acids can enable precise temporal control of proteins. Light-responsive groups on unnatural amnio acids have been used to activate or inhibit proteins as well as to reversibly switch on or off the catalytic function. For example, the group of Professor Peter Schultz have introduced DMNB-caged-Serine
Unnatural amino acids have also been used effectively in proteomics studies. Incorporating a photoactivatable residue such as AbK (Cat. No. 5113) into the protein of interest enables protein interactions in the native intracellular environment to be captured. Another important demonstration of optical control of protein function is with photoswitchable amino acids. These compounds typically include an azobenzene chromophore and undergo reversible cis-trans isomerization upon excitation. This technique has been used successfully in the laboratory of Professor Sterner to study the effect of allosteric regulation on imidazole glycerol phosphate synthase (ImGPS) enzyme activity. (Reference: Kneuttinger et al. (2019) Light regulation of enzyme allostery through photo-responsive unnatural amino acids. 26 1501 PMID 31495713
Advantages and Disadvantages of Optical-Controlled Genetic code expansion:
- The speed at which optical-controlled genetic code expansion can be manipulated is milliseconds to seconds, versus the minute to hour time scale of translational control.
- Optical-controlled genetic code expansion provides precise spatial control, allowing detailed analysis of subcellular compartments.
- UVA light has been shown to alter some cellular activity and may not be suitable to all experimental types.
Genetic Code Expansion: Chemical Control of Unnatural Amino Acid Function
Small molecules can be used to activate or inhibit proteins through their action on unnatural amino acids. For example, removal of protecting groups on an appropriately incorporated unnatural amino acid has been shown to be a useful method for switching on protein function. Chemical control of unnatural amino acids function has also been used to deactivate enzymes. Through bioorthogonal tethering of a small molecule to an unnatural amino acid containing a reactive handle, proximity induced inhibition can be achieved. This approach has been used successfully to inhibit an intracellular kinase with no known inhibitor. (Reference: Tsai et al. (2015) Selective, rapid and optically switchable regulation of protein function in live mammalian cells. 7 554. PMID 26100803
A major advantage of small molecule controlled genetic code expansion is that it can be used in vivo. It also has good temporal control, with faster responses than translational control, but slower than optical control.
| Control Type | Temporal Control | Spatial control | Reversable |
|---|---|---|---|
| Translational Control | Slow | Yes, when administered locally | Yes, but slow |
| Optical Control | Fast | Excellent, subcellular levels | Possible with photoswitchable amino acids |
| Chemical Control | Moderate speed | Yes, when administered locally | NA |
Table 1: Advantages and disadvantages of protein functional control approaches enabled by genetic code expansion. Adapted from Nodling, et al (2019) Using genetically incorporated unnatural amino acids to control protein functions in mammalian cells. Essays Biochem. 63 237 PMID 31092687
Using Fluorescent D-Amino Acids (FDAAs) to Image Bacteria
Fluorescently-labeled wheat germ agglutinin (FWGA) and fluorescently-labeled antibiotics, have been used as molecular probes to study peptidoglycans (PGs). However, both of these probes have serious drawbacks; bacterial growth is impeded by fluorescent antibiotics, and FWGA cannot cross gram-negative bacteria cell walls.
Fluorescent D-amino acids (FDAAs) are metabolic probes that take advantage of the promiscuous nature of the bacterial PG 60 biosynthetic pathway. FDAAs are small molecules conjugated to D-amino acids, which are incorporated into new polyglycogen chains as bacteria grow and proliferate. They efficiently label peptidoglycans in bacterial cell walls in situ, which enables the investigation of bacterial physiology, as well as bacterial growth. FDAAs can be used in both gram-negative and gram-positive bacterial species, in real-time and in live cells, with minimal toxic effects. They are suitable probes for use with both confocal and super-resolution microscopy.
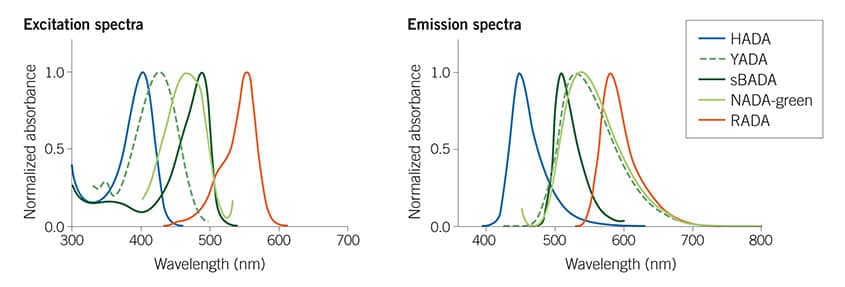
Figure 1: The Tocris Fluorescent D-amino Acids (FDAAs) collection spans the visible spectrum, from blue HADA (#6647) to red RADA (#6649) providing many options for multiplexing experiments. For more information see Hsu et al (2017) Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem.Sci. 8 6313 PMID: 28989665
Literature for Unnatural Amino Acids / Non Canonical Amino Acids
Tocris offers the following scientific literature for Unnatural Amino Acids / Non Canonical Amino Acids to showcase our products. We invite you to request* your copy today!
*Please note that Tocris will only send literature to established scientific business / institute addresses.
Fluorescent Dyes and Probes Research Product GuideUpdated
This product guide provides a background to the use of Fluorescent Dyes and Probes, as well as a comprehensive list of our:
- Fluorescent Dyes, including Janelia Fluor® Dyes
- Fluorescent Probes for Advanced Microscopy and Live Cell Imaging
- Fluorescent Probes to Support In Vivo, Deep Tissue, and Bioluminescence Imaging
- Fluorescent Probes and Reagents for Organoids and 3D Cell Culture Imaging
- Aptamer-based RNA Imaging Reagents
- Fluorescent Probes for Imaging Bacteria
- TSA VividTM Fluorophore Kits
- TSA Reagents for Enhancing IHC, ICC & FISH Signals
- Tissue Clearing Kits and Reagents
